SIGNAL: A Population-Scale Resource of Germline Variants and Accompanying Somatic Alterations in Cancer
We propose to build a public web resource that hosts the prevalence data of germline variants in dozens of cancer types as well as their companying somatic alterations in the same genes in the tumor. It will fill the critical gap of a public database for the dissemination and interpretation of germline variants in cancer and has the potential to become a critical reference resource for the cancer research and clinical genetics communities.
Learn More About Their Work:
Nature Magazine, Vol. 53 Issue 11, Nov. 5, 2021: The context-specific role of germline pathogenicity in tumorigenesis
Final Summary of Work
May 3, 2022
Original Aims
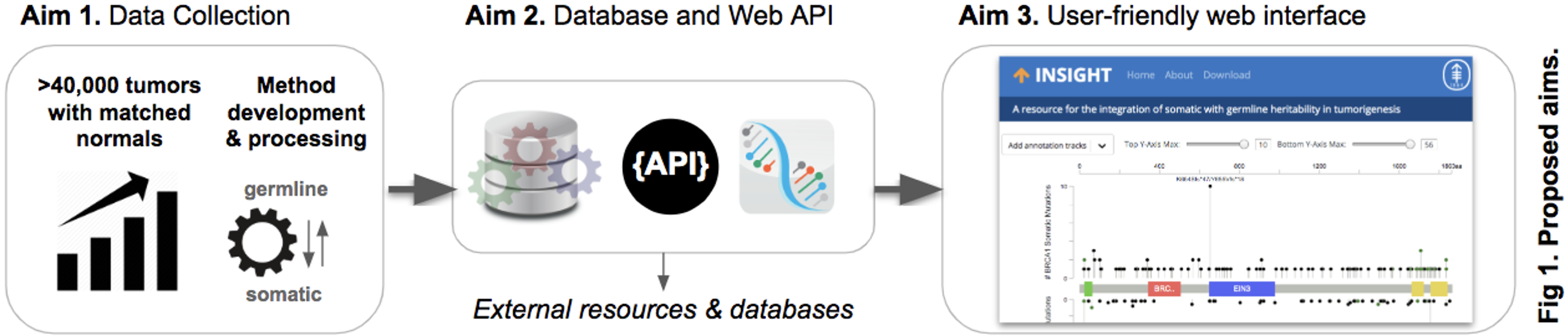
To improve our understanding of the prevalence of specific germline mutations across different cancer types and the rates of somatic changes in the accompanying tumors, we propose to build a public web resource that hosts the prevalence data of germline variants in dozens of cancer types as well as their companying somatic alterations in the same genes in the tumor. It will fill the critical gap of a public database for the dissemination and interpretation of germline variants in cancer and has the potential to become a critical reference resource for cancer research and clinical genetics communities. Specifically, we proposed to build a data pipeline that periodically re-processes and releases data from our growing clinical sequencing cohort (Aim 1), store the data in a publicly available database with a web API (Aim 2), and disseminate the data to the broader cancer research community via a user-friendly website (Aim 3) (Fig. 1). Through these aims, we will establish a central, fundamental one-of-a-kind reference dataset and public portal to catalyze future studies and aid the interpretation of germline variants of uncertain significance.
Summary of Work
Over the duration of the award, we have accomplished all of our aims, and SIGNAL is operational and publicly available at https://signaldb.org with an intuitive user interface (UI) and a programing application interface (API). Through its API, the SIGNAL data has also been integrated into cBioPortal (https://cbioportal.org) assisting thousands of users to interpret mutations. All code for the project is available via Github at https://github.com/knowledgesystems/signal/. Specifically, we have accomplished the following tasks, grouped by the aim:
Aim 1. Collection and processing of germline and somatic variant data in cancer.
We have built a data pipeline to a) collect patient-level clinical data (cancer type, race, gender, and binned age) and genomics data (germline and somatic variants) from the MSK-IMPACT cohort; b) predict pathogenicity and penetrance stratification for each germline variant; and c) generate summary data including prevalences of each somatic and germline mutations across and within cancer types, frequencies of somatic biallelic inactivation accompanying each germline variant, and distributions of clinical features (e.g., age, MSI score, TMB, HRD LST, etc) for each germline variant. Data of b & c from the first 17,152 MSK patients by Srinivasan et al have been released through SIGNAL web API (Aim 2) and website (Aim 3). Subsequent iterations of this pipeline have been used to analyze a combined set of >51,000 patients receiving MSK-IMPACT testing, the results of which are currently undergoing manual quality control.
Aim 2. Implementation of database and web API.
To facilitate data sharing and programmatic access, the data are hosted in a public database that is accessible through a web application programming interface (API). We have utilized the database and API architecture of an existing platform, Genome Nexus. All data in Aim 1 were imported into its Mongo database, and new endpoints were developed to allow query and access to the data on different levels (cohort, gene, and variant). See internal endpoint signal-mutation-controller at https://www.genomenexus.org/swagger-ui.html.
Aim 3. Development of a user-friendly web interface.
In addition to the API and client libraries, we have also developed a user-friendly and open-access website (https://signaldb.org), which allows researchers to a) study the landscape of germline variants and their interplay with somatic variants across cancer types or within a particular cancer type; b) visualize and analyze germline and somatic variants in individual genes; c) search and interpret biological and clinical relevance of individual germline variants in cancer patients. The website has served over 1,000 users in the past year.
In summary, SIGNAL is the first database that provides integrated summary data of germline and somatic variants in the cancer patient population. It is also the first resource that provides the rates of zygosity alterations affecting germline alleles in the associated tumors. It has the potential to become a critical and widely-utilized reference resource for the cancer community, facilitating the study of cancer germline mutations, their interplay with somatic alterations, and their clinical characterization and interpretation when observed in cancer patients.